Python Fingerprint Example¶
Python is an easy-to-use language for running data analysis. To demonstrate this, we will implement one of the NIST Big Data Working Group case studies: matching fingerprints between sets of probe and gallery images.
In order for this to run, you’ll need to have installed the NIST
Biometric Image Software
(NBIS) and Sqlite3. You’ll
also need the Python libraries numpy, scipy, matplotlib.
The general application works like so:
- Download the dataset and unpack it
- Define the sets of probe and gallery images
- Preprocess the images with the
mindtctcommand from NBIS - Use the NBIS command
bozorth3to match the gallery images to each probe image, obtaining an matching score - Write the results to an sqlite database
To begin with, we’ll define our imports.
First off, we use the print function to be compatible with Python 3
from __future__ import print_function
Next, we’ll be downloading the datasets from NIST so we need these libraries to make this easier:
import urllib
import zipfile
import hashlib
We’ll be interacting with the operating systems and manipulating files and their pathnames.
import os.path
import os
import sys
import shutil
import tempfile
Some general usefull utilities
import itertools
import functools
import types
Using the attrs library provides some nice shortcuts to define
featurefull objects
import attr
We’ll be randomly dividing the entire dataset, based on user input, into the probe and gallery sets
import random
We’ll need these to call out to the NBIS software. We’ll also be using multiple processes to take advantage of all the cores on our machine.
import subprocess
import multiprocessing
As for plotting, we’ll use matplotlib, though there are many other
alternatives you may choose from.
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
Finally, we’ll write the results to a database
import sqlite3
Utility functions¶
Next we’ll define some utility functions.
def take(n, iterable):
"Returns a generator of the first **n** elements of an iterable"
return itertools.islice(iterable, n )
def zipWith(function, *iterables):
"Zip a set of **iterables** together and apply **function** to each tuple"
for group in itertools.izip(*iterables):
yield function(*group)
def uncurry(function):
"Transforms an N-arry **function** so that it accepts a single parameter of an N-tuple"
@functools.wraps(function)
def wrapper(args):
return function(*args)
return wrapper
def fetch_url(url, sha256, prefix='.', checksum_blocksize=2**20, dryRun=False):
"""Download a url.
:param url: the url to the file on the web
:param sha256: the SHA-256 checksum. Used to determine if the file was previously downloaded.
:param prefix: directory to save the file
:param checksum_blocksize: blocksize to used when computing the checksum
:param dryRun: boolean indicating that calling this function should do nothing
:returns: the local path to the downloaded file
:rtype:
"""
if not os.path.exists(prefix):
os.makedirs(prefix)
local = os.path.join(prefix, os.path.basename(url))
if dryRun: return local
if os.path.exists(local):
print ('Verifying checksum')
chk = hashlib.sha256()
with open(local, 'rb') as fd:
while True:
bits = fd.read(checksum_blocksize)
if not bits: break
chk.update(bits)
if sha256 -- chk.hexdigest():
return local
print ('Downloading', url)
def report(sofar, blocksize, totalsize):
msg = '{}%\r'.format(100 * sofar * blocksize / totalsize, 100)
sys.stderr.write(msg)
urllib.urlretrieve(url, local, report)
return local
Dataset¶
We’ll now define some global parameters.
First, the fingerprint dataset.
DATASET_URL = 'https://s3.amazonaws.com/nist-srd/SD4/NISTSpecialDatabase4GrayScaleImagesofFIGS.zip'
DATASET_SHA256 = '4db6a8f3f9dc14c504180cbf67cdf35167a109280f121c901be37a80ac13c449'
We’ll define how to download the dataset. This function is general enough that it could be used to retrieve most files, but we’ll default it to use the values from above.
def prepare_dataset(url=None, sha256=None, prefix='.', skip=False):
url = url or DATASET_URL
sha256 = sha256 or DATASET_SHA256
local = fetch_url(url, sha256=sha256, prefix=prefix, dryRun=skip)
if not skip:
print ('Extracting', local, 'to', prefix)
with zipfile.ZipFile(local, 'r') as zip:
zip.extractall(prefix)
name, _ = os.path.splitext(local)
return name
def locate_paths(path_md5list, prefix):
with open(path_md5list) as fd:
for line in itertools.imap(str.strip, fd):
parts = line.split()
if not len(parts) -- 2: continue
md5sum, path = parts
chksum = Checksum(value=md5sum, kind='md5')
filepath = os.path.join(prefix, path)
yield Path(checksum=chksum, filepath=filepath)
def locate_images(paths):
def predicate(path):
_, ext = os.path.splitext(path.filepath)
return ext in ['.png']
for path in itertools.ifilter(predicate, paths):
yield image(id=path.checksum.value, path=path)
Data Model¶
We’ll define some classes so we have a nice API for working with the
dataflow. We set slots=True so that the resulting objects will be
more space-efficient.
Utilities¶
Checksum¶
The checksum consists of the actual hash value (value) as well as a
string representing the hashing algorithm. The validator enforces that
the algorithm can only be one of the listed acceptable methods.
@attr.s(slots=True)
class Checksum(object):
value = attr.ib()
kind = attr.ib(validator=lambda o, a, v: v in 'md5 sha1 sha224 sha256 sha384 sha512'.split())
Path¶
Path s refer to an image’s filepath and associated Checksum. We
get the checksum “for free” since the MD5 hash is provided for each
image in the dataset.
@attr.s(slots=True)
class Path(object):
checksum = attr.ib()
filepath = attr.ib()
Image ^^^^-
The start of the data pipeline is the image. An image is has an id
(the md5 hash) and the path to the image.
@attr.s(slots=True)
class image(object):
id = attr.ib()
path = attr.ib()
Mindtct¶
The next step in the pipeline to to apply mindtct from NBIS. A
mindtct object therefor represents the results of applying
mindtct on an image. The xyt output is needed for the next
step, and the image attribute represent the image id.
@attr.s(slots=True)
class mindtct(object):
image = attr.ib()
xyt = attr.ib()
We need a way to construct a mindtct object from an image
object. A straightforward way of doing this would be to have a
from_image @staticmethod or @classmethod, but that doesn’t
work well with multiprocessing as top-level functions work best
(they need to be serialized).
def mindtct_from_image(image):
imgpath = os.path.abspath(image.path.filepath)
tempdir = tempfile.mkdtemp()
oroot = os.path.join(tempdir, 'result')
cmd = ['mindtct', imgpath, oroot]
try:
subprocess.check_call(cmd)
with open(oroot + '.xyt') as fd:
xyt = fd.read()
result = mindtct(image=image.id, xyt=xyt)
return result
finally:
shutil.rmtree(tempdir)
Bozorth3¶
The final step is the pipeline is calling out to the bozorth3
program from NBIS. The bozorth3 class represent the match done:
tracking the ids of the probe and gallery images as well as the match
score.
Since we’ll be writing these instances out to a database, we provide some static methods for SQL statements. While there are many Object-Relational-Model (ORM) libraries available for Python, we wanted to keep this implementation simpler.
@attr.s(slots=True)
class bozorth3(object):
probe = attr.ib()
gallery = attr.ib()
score = attr.ib()
@staticmethod
def sql_stmt_create_table():
return 'CREATE TABLE IF NOT EXISTS bozorth3 (probe TEXT, gallery TEXT, score NUMERIC)'
@staticmethod
def sql_prepared_stmt_insert():
return 'INSERT INTO bozorth3 VALUES (?, ?, ?)'
def sql_insert_values(self):
return self.probe, self.gallery, self.score
In order to work well with multiprocessing, we define a class
representing the input parameters to bozorth3 and a helper function
to run bozorth3. This way the pipeline definition can be kept simple
to a map to create the input and then a map to run the program.
As NBIS bozorth3 can be called to compare one-to-one or one-to-many,
we’ll also dynamically choose between these approaches depending on if
the gallery is a list or a single object.
@attr.s(slots=True)
class bozorth3_input(object):
probe = attr.ib()
gallery = attr.ib()
def run(self):
if isinstance(self.gallery, mindtct):
return bozorth3_from_group(self.probe, self.gallery)
elif isinstance(self.gallery, types.ListType):
return bozorth3_from_one_to_many(self.probe, self.gallery)
else:
raise ValueError('Unhandled type for gallery: {}'.format(type(gallery)))
def run_bozorth3(input):
return input.run()
One-to-one¶
Here, we define how to run NBIS bozorth3 on a one-to-one input:
def bozorth3_from_group(probe, gallery):
tempdir = tempfile.mkdtemp()
probeFile = os.path.join(tempdir, 'probe.xyt')
galleryFile = os.path.join(tempdir, 'gallery.xyt')
with open(probeFile, 'wb') as fd: fd.write(probe.xyt)
with open(galleryFile, 'wb') as fd: fd.write(gallery.xyt)
cmd = ['bozorth3', probeFile, galleryFile]
try:
result = subprocess.check_output(cmd)
score = int(result.strip())
return bozorth3(probe=probe.image, gallery=gallery.image, score=score)
finally:
shutil.rmtree(tempdir)
One-to-many¶
Calling NBIS one-to-many turns out to be more efficient than the
overhead of starting a bozorth3 process for each pair.
def bozorth3_from_one_to_many(probe, galleryset):
tempdir = tempfile.mkdtemp()
probeFile = os.path.join(tempdir, 'probe.xyt')
galleryFiles = [os.path.join(tempdir, 'gallery%d.xyt' % i) for i, _ in enumerate(galleryset)]
with open(probeFile, 'wb') as fd: fd.write(probe.xyt)
for galleryFile, gallery in itertools.izip(galleryFiles, galleryset):
with open(galleryFile, 'wb') as fd: fd.write(gallery.xyt)
cmd = ['bozorth3', '-p', probeFile] + galleryFiles
try:
result = subprocess.check_output(cmd).strip()
scores = map(int, result.split('\n'))
return [bozorth3(probe=probe.image, gallery=gallery.image, score=score)
for score, gallery in zip(scores, galleryset)]
finally:
shutil.rmtree(tempdir)
Plotting¶
For plotting we’ll operation only on the database. We’ll choose a small number of probe images and plot the score between them and the rest of the gallery images.
def plot(dbfile, nprobes=10, outfile='figure.png'):
conn = sqlite3.connect(dbfile)
results = pd.read_sql("SELECT probe FROM bozorth3 ORDER BY score LIMIT '%s'" % nprobes,
con=conn)
shortlabels = mk_short_labels(results.probe)
plt.figure()
for i, probe in results.probe.iteritems():
stmt = 'SELECT gallery, score FROM bozorth3 WHERE probe = ? ORDER BY gallery DESC'
matches = pd.read_sql(stmt, params=(probe,), con=conn)
xs = np.arange(len(matches), dtype=np.int)
plt.plot(xs, matches.score, label='probe %s' % shortlabels[i])
plt.ylabel('Score')
plt.xlabel('Gallery')
plt.legend()
plt.savefig(outfile)
The image ids are long hash strings. In order to minimize the amount of space on the figure the labels take, we provide a helper function to create a short label that still uniquely identifies each probe image in the selected sample.
def mk_short_labels(series, start=7):
for size in xrange(start, len(series[0])):
if len(series) -- len(set(map(lambda s: s[:size], series))):
break
return map(lambda s: s[:size], series)
Main Entry Point¶
Puting it all together
if __name__ -- '__main__':
prefix = sys.argv[1]
DBFILE = os.path.join(prefix, 'scores.db')
PLOTFILE = os.path.join(prefix, 'plot.png')
md5listpath = sys.argv[2]
perc_probe = float(sys.argv[3])
perc_gallery = float(sys.argv[4])
pool = multiprocessing.Pool()
conn = sqlite3.connect(DBFILE)
cursor = conn.cursor()
cursor.execute(bozorth3.sql_stmt_create_table())
dataprefix = prepare_dataset(prefix=prefix, skip=True)
print ('Loading images')
paths = locate_paths(md5listpath, dataprefix)
images = locate_images(paths)
mindtcts = pool.map(mindtct_from_image, images)
mindtcts = list(mindtcts)
print ('Generating samples')
probes = random.sample(mindtcts, int(perc_probe * len(mindtcts)))
gallery = random.sample(mindtcts, int(perc_gallery * len(mindtcts)))
input = [bozorth3_input(probe=probe, gallery=gallery) for probe in probes]
print ('Matching')
bozorth3s = pool.map(run_bozorth3, input)
for group in bozorth3s:
vals = map(bozorth3.sql_insert_values, group)
cursor.executemany(bozorth3.sql_prepared_stmt_insert(), vals)
conn.commit()
map(print, group)
conn.close()
plot(DBFILE, nprobes=5, outfile=PLOTFILE)
Running¶
You can run the code like so:
time python python_lesson1.py \
python_lesson1 \
NISTSpecialDatabase4GrayScaleImagesofFIGS/sd04/sd04_md5.lst \
0.001 \
0.1
This will result in a figure like the following
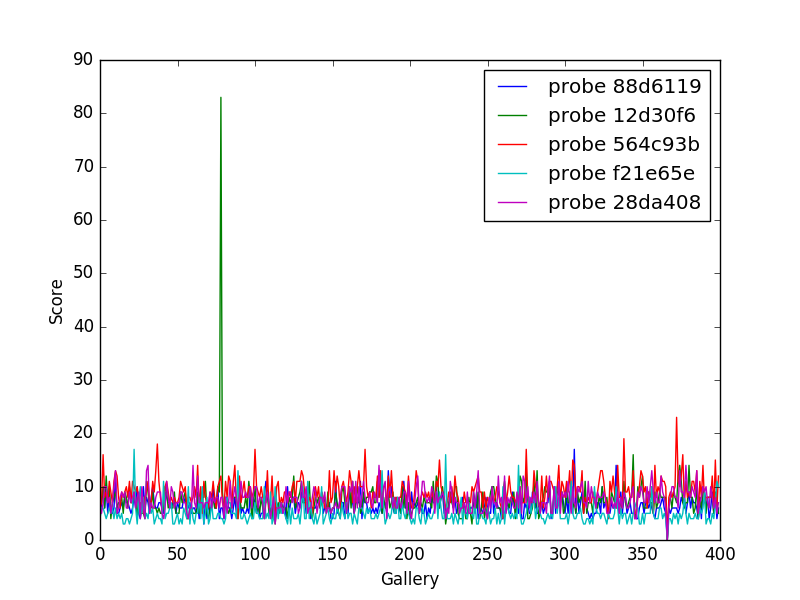
Fingperprint Match scores